Why use Germline Cancer Evidence Base?
Genetic mutations in specific genes elevate the risk of developing cancer, collectively referred to as hereditary cancer syndromes. Understanding associations of genetic mutations with cancer syndrome helps tailor diagnostic and preventive measures for carriers of mutations. Classification of variants identified by a typical sequencing or panel assay requires one or more types of evidence. A key source of evidence that helps support variant classification is published articles. For example, at least six evidence codes (PP1, PM6, PS2, PP4, BS4, and BP3) described in ACMG guidelines use published case reports or series in one or another form to support variant classification. However, the enormous quantity of published literature (with more than 30 million articles) makes finding the relevant article difficult. Experts-led curation efforts help collect, curate, and compile data for use in variation classification. However, such efforts are expensive, and the scale is limited. Nucleati is on a mission to identify essential components and automate relevant literature collection and curation processes. Nucleati germline cancer evidence base is an output of a fully automated data curation pipeline. The pipeline collects, curates, and normalizes data for knowledge discovery.
The germline cancer evidence base currently comprises knowledge across two domains: case reports and case series. Case reports include a detailed description of one patient, and cancer series are outcomes of studies on multiple patients.
How did we create the Nucleati germline cancer evidence base?
The pipeline consists of three modules that go through more than 30 million articles and produce a few thousand structured and ontology mapped documents.
Text classification model
There are nearly 31 million articles available in NCBI PubMed. The number of articles reporting a patient(s) with germline mutation who developed cancers is few thousand. The first step of knowledge discovery is finding this relatively negligible number of articles from PubMed programmatically. To do so, we generate a training set for every set of articles of interest and train AI models to identify them correctly. The developed model scans all abstracts available in PubMed to identify relevant articles. A typical model takes 400–500 articles of interest and 10–20k background articles.
Entity recognition models
Using the transfer learning principles, a general-purpose language model, BERT, is refined using a training set for entity recognition developed by Nucleati. Unlike general-purpose entity recognition models, the models developed in this manner are domain experts. Because the purpose is to identify relevant entities from a focused set of articles, they efficiently recognize them compared to general-purpose entity recognition models.
Ontology mapping, validation, and term normalization
Unless the identified entities are mapped to an ontology or normalized, the identified terms carry little meaning and hamper filtering data. Nucleati uses a meta thesaurus for mapping terms to ontology. For entities that are irrelevant for ontology mapping, proprietary methods developed by Nucleati are used to validate and normalize various entities.
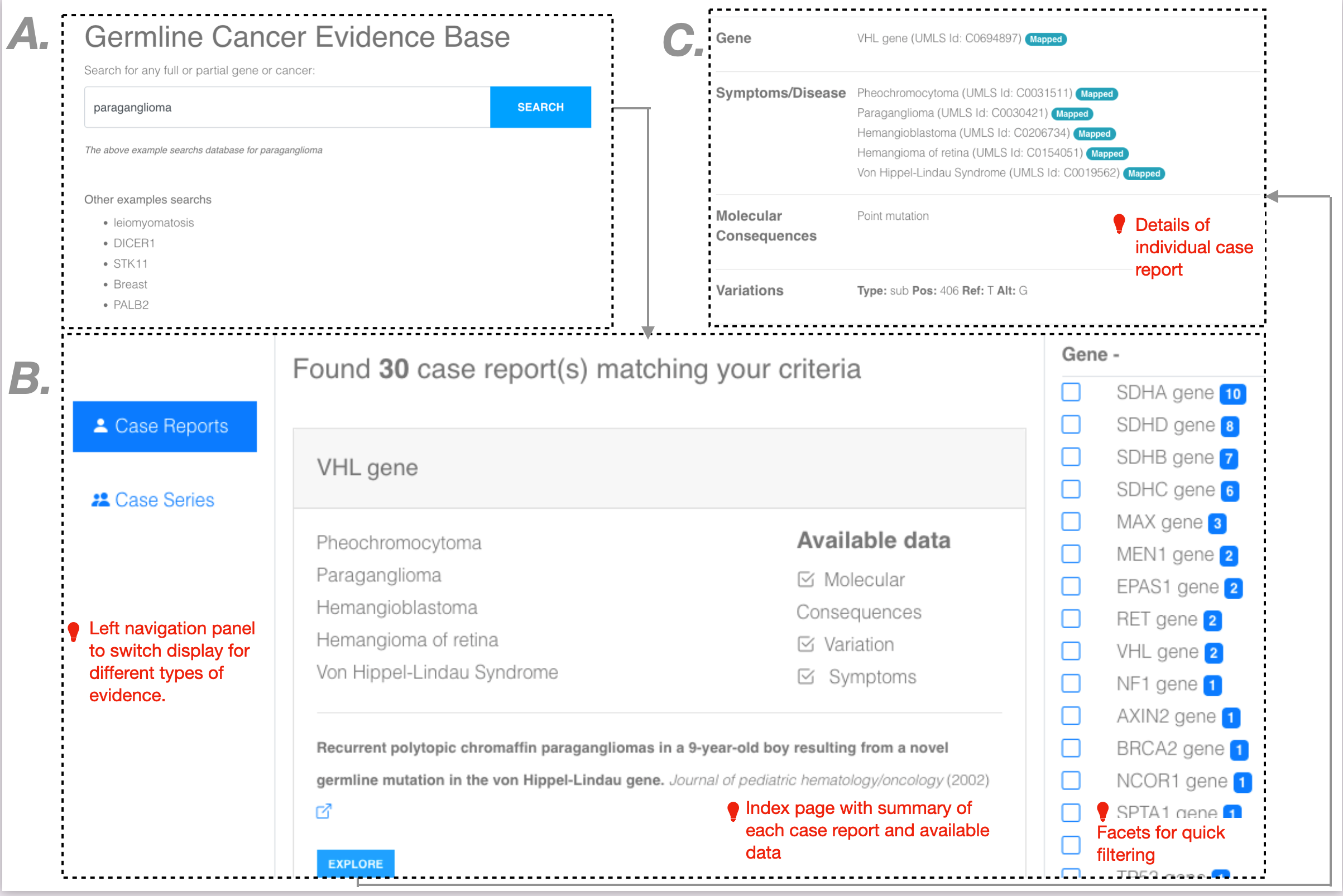
The user interface for browsing collected evidence:
The following snapshot describes various UI components of the germline cancer evidence base. Broadly, the UI consists of three main parts: (1) search page, (2) index page, and (3) details of individual AI curated reports. The search page supports searching the database using full or partial genes or cancer. The middle panel of the index page summarizes all hits for a given category of data. The left navigation panel provides a way to switch between different data categories. Based on the data category, the index page provides an easy way to filter the retrieved hits using facets. In addition to providing a basic summary, the index page also notes what type of data is collected using the AI. More details of each identified hit are available through the “Explore” button associated with each result. More details are available only to authenticated users. All the data is freely available to the user.
Summary and future outlook
Nucleati aims to continue efforts to develop and make AI-curated data available in the field where it is most necessary. One of the immediate interests is identifying and curating case studies, case series, case-control data in genetic medicine, and therapeutic interventions such as CART/Fecal transplantation. We also plan to collect and use AI to curate data for evidence for pediatric diseases and interventions.